Advances in Tumor Modeling Techniques Could Revolutionize Cancer Treatment
Cancer remains one of the most formidable challenges in modern medicine, characterized by its genetic diversity, complex molecular profiles, and intricate cellular compositions. As researchers continue to explore ways to develop effective therapies, understanding the full complexity of human cancers has proven to be a critical hurdle. Traditional models such as two-dimensional cell cultures and animal testing have struggled to capture the nuanced interactions within the tumor microenvironment, particularly in solid tumors.
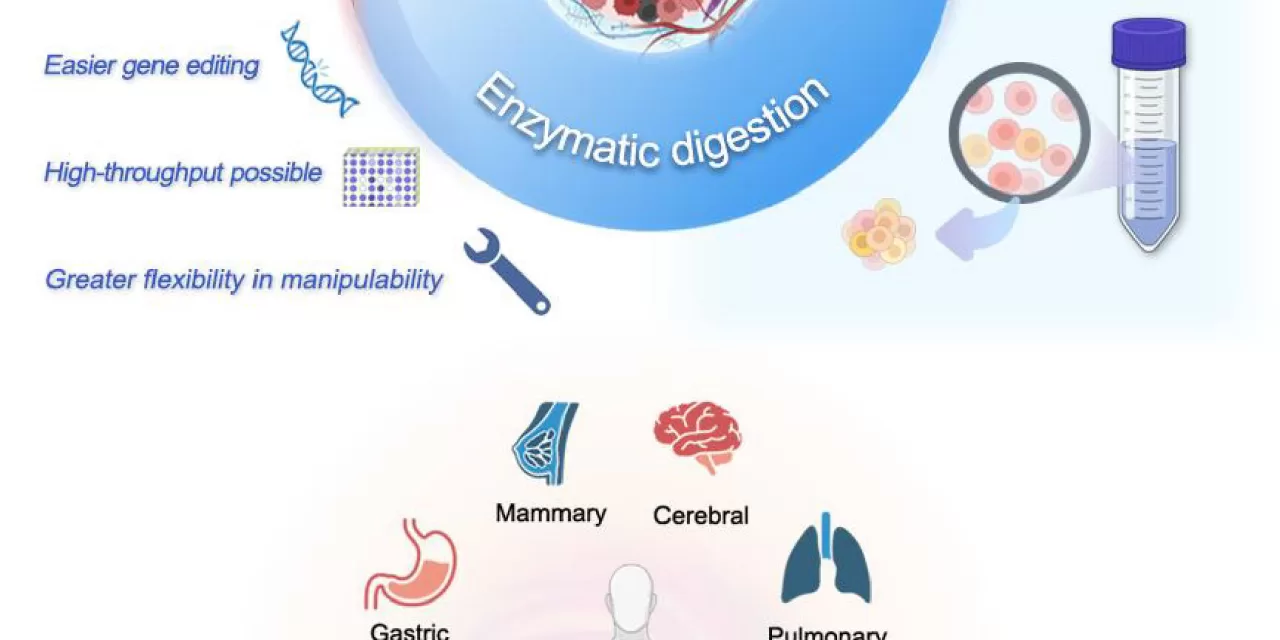
A groundbreaking study by researchers from Henan Provincial People’s Hospital and the Academy of Medical Science has shed new light on the methods used to dissociate solid tumors for use in cancer models. This research explores two key dissociation techniques—mechanical dissociation and enzymatic digestion—to create patient-derived organoids (PDOs), offering vital insights into which method best preserves the tumor’s characteristics for research purposes.
Mechanical vs. Enzymatic Dissociation
The study, recently published in Cell Organoid, reveals that mechanical dissociation, which involves physically breaking down tissue, is more effective at maintaining the integrity of the tumor microenvironment. This method preserves the three-dimensional structure and cellular heterogeneity of the tumor, critical elements for accurately modeling human cancers and understanding their behavior.
On the other hand, enzymatic digestion—often used for its ability to break down extracellular matrices more efficiently—results in a more homogeneous cell population. While this makes it ideal for large-scale drug screening due to its reproducibility and controllability, it sacrifices the complex tissue interactions present in the native tumor.
Organoid Properties and Implications for Personalized Medicine
One of the most significant takeaways from the study is the impact of dissociation methods on organoid properties such as stemness, differentiation potential, and tumor heterogeneity. These factors are key to developing personalized cancer treatments, as they help researchers better understand how tumors evolve, how they develop resistance to drugs, and how they metastasize.
According to Dr. Haijun Li, the corresponding author of the study, the findings provide essential guidance to researchers on how to choose the best dissociation technique based on their specific research goals. “This article offers new insights into the complexities of mechanical and enzymatic dissociation methods, helping to improve the accuracy of organoid-based cancer models and contributing to the development of more effective personalized treatments,” Dr. Li stated.
Future of Cancer Research and Therapy
By refining tissue dissociation methods, the research team believes they are paving the way for more precise cancer models, which could transform the way drug screening is conducted. More accurate models will allow for better identification of therapeutic targets, potentially leading to more effective treatments tailored to individual tumor characteristics.
As organoid-based cancer models continue to evolve, this study promises to make a lasting impact on oncology research, providing the foundation for developing therapies that are not only more targeted but also capable of addressing the complex nature of cancer.
This advancement in cancer research could have profound implications for patient outcomes, offering hope for personalized treatments that better align with the genetic and phenotypic characteristics of each patient’s tumor.
Further Reading: Jing Ren et al, “The Pros and Cons of Mechanical Dissociation and Enzymatic Digestion in Patient-Derived Organoid Cultures for Solid Tumor,” Cell Organoid (2024). DOI: 10.26599/CO.2024.9410009