January 24, 2025 – EMBL Grenoble, France
In a breakthrough study, researchers from the Kowalinski group at the European Molecular Biology Laboratory (EMBL) in Grenoble have uncovered vital structural details of a critical protein complex in Trypanosoma brucei, the parasite responsible for causing devastating diseases like Human African Trypanosomiasis (sleeping sickness), Chagas disease, and Nagana in cattle. These diseases, classified as neglected tropical diseases (NTDs), continue to affect millions globally, despite substantial progress in combating sleeping sickness.
While the World Health Organization (WHO) has made considerable strides in reducing sleeping sickness cases, Chagas disease remains a persistent challenge. No effective vaccine currently exists, and treatment options are complex and often inadequate. With climate change poised to expand the range of insect-borne diseases, understanding the biology of trypanosomes has become increasingly urgent.
Unraveling the Mysteries of mRNA Processing
Like all eukaryotic organisms, trypanosomes use messenger RNA (mRNA) to carry genetic instructions for protein synthesis. These instructions are crucial for the parasite’s survival, especially during the infection process. In humans and trypanosomes, mRNA undergoes extensive processing before it can be translated into proteins. However, significant differences exist between the two organisms in how mRNAs are processed. Understanding these differences could lead to the development of drugs that specifically target the parasites without affecting human cells.
In their recent publication in Nature Communications, the Kowalinski group focused on the nuclear cap-binding complex in T. brucei. This complex binds to all mRNAs in the parasite and is essential for proper mRNA processing, making it a potential drug target.
A Structural Breakthrough
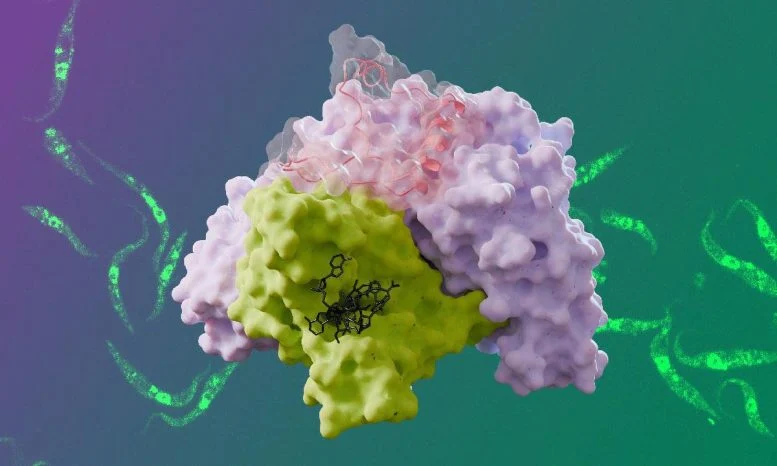
The research revealed key differences between the trypanosomal cap-binding complex and its human counterpart, which consists of two subunits. In contrast, the trypanosomal version contains four subunits, with three of them previously not well understood. Using cryo-electron microscopy (cryo-EM) and small-angle X-ray scattering, the team was able to explore the complex’s structure in detail, revealing that it consists of two lobes. One lobe closely resembles the human complex, while the second lobe is connected by a flexible protein.
“The flexibility of this complex presented a challenge in our research,” explained Harald Bernhard, the study’s lead author. “Cryo-EM alone couldn’t resolve the flexible regions, so we turned to small-angle X-ray scattering at the European Synchrotron Radiation Facility.”
Additionally, the team discovered that T. brucei mRNAs possess a highly modified RNA cap structure, which differs from the caps found in other organisms. This unique cap structure provides insight into how the cap-binding complex interacts with RNA, highlighting a potential second binding site for RNA within the complex.
Implications for Drug Development
Given the importance of the cap-binding complex in the survival of the parasite and its structural differences from humans, it presents a promising target for drug development. “This avenue is currently being explored by a PhD student in my group,” said Eva Kowalinski, the principal investigator of the study.
Furthermore, the research will contribute to a broader understanding of RNA processing in T. brucei, which is also crucial in the study of eukaryotic life and evolution. Kowalinski’s group has also received funding from the European Research Council (ERC) to explore trans-splicing mechanisms in trypanosomes, an innovative approach to RNA editing that holds potential for gene therapy.
Future Prospects
While this research marks a significant step forward, there is still much to be done before this discovery leads to therapeutic applications. The data will be instrumental in guiding future studies aimed at developing targeted drugs for diseases like sleeping sickness and Chagas, which continue to devastate vulnerable populations.
The Kowalinski group’s work shines a light on a critical aspect of parasite biology and offers hope for the future development of more effective treatments for neglected tropical diseases.
Disclaimer: The findings discussed in this article are based on the study “Structural basis of Spliced Leader RNA recognition by the Trypanosoma brucei cap-binding complex,” published in Nature Communications (January 15, 2025), and are subject to further validation and research.
For more details, visit Nature Communications.