In a landscape increasingly focused on developing new cancer therapies, scientists are rediscovering the potential of old drugs. One such drug, hydroxychloroquine—originally an anti-malarial medication—has been repurposed in the fight against cancer. While it shows promise in blocking essential resources to cancer cells, recent clinical trials have yielded disappointing results. The reason? Cancer cells often develop resistance to hydroxychloroquine.
A research team led by Dr. Joe Delaney at the Medical University of South Carolina Hollings Cancer Center has made significant strides in understanding this resistance. Their findings, published in Cell Cycle, reveal that resistance to hydroxychloroquine does not occur by reinstating cancer cells’ recycling capabilities as previously thought. Instead, resistance emerges through alterations in cancer cells’ division, metabolism, and export pathways.
The Promise of Repurposing Drugs
The concept of repurposing existing medications for new treatments is not novel. For example, aspirin transitioned from a pain reliever to a widely-used blood thinner, while thalidomide has found new life as a treatment for certain cancers and leprosy. As cancer therapies evolve to target specific proteins, researchers like Delaney advocate for revisiting older drugs that demonstrate broader, multi-target effects.
“Targeting single proteins can be extremely effective, but specificity can lead to resistance,” Delaney explained. He likens the process to securing a hotel hallway, where targeting one door may prevent entry, but cancer will inevitably find another way in. Older drugs like hydroxychloroquine may act on multiple targets, making it more challenging for cancer cells to adapt.
Originally employed to treat malaria, hydroxychloroquine’s potential in oncology was explored in the mid-2000s due to its ability to inhibit autophagy—a cellular process crucial for cancer cell survival. By blocking autophagy, the drug aims to starve cancer cells of the resources they need to thrive.
Despite its initial promise, many clinical trials have produced lackluster results. “What we don’t know is why so many of these clinical trials have failed,” said Delaney. “We’re trying to figure out why hydroxychloroquine works or doesn’t work in certain situations in cancer.”
New Insights into Resistance Mechanisms
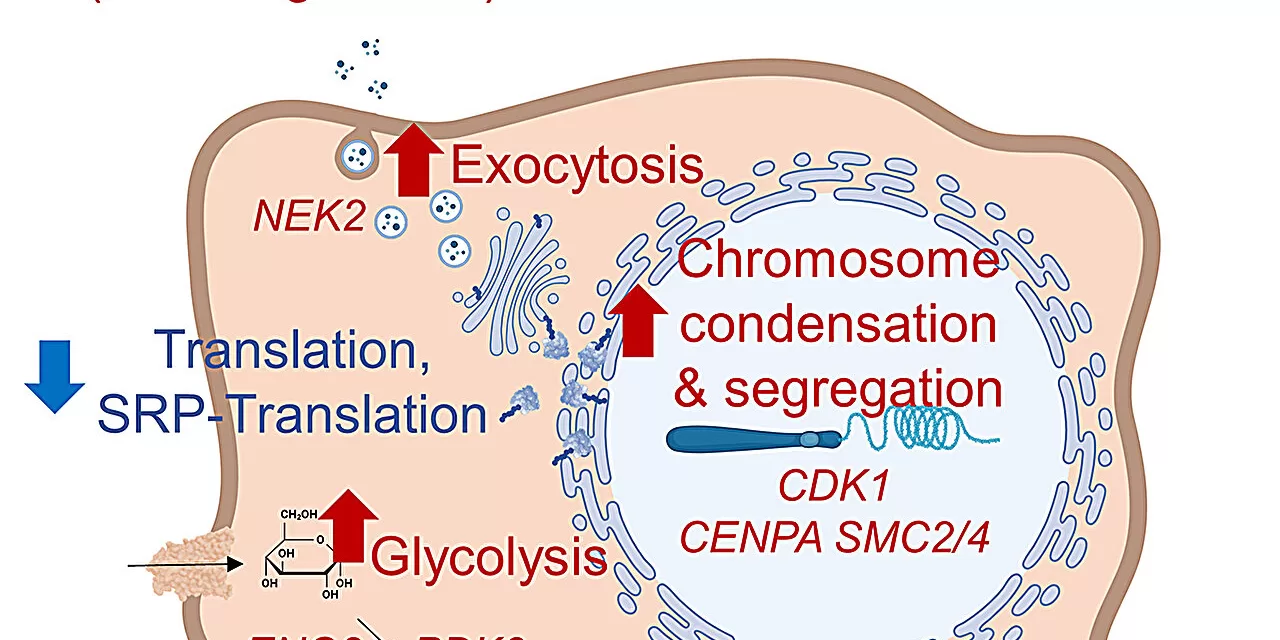
To tackle these questions, Delaney’s team conducted an extensive investigation into hydroxychloroquine’s impact on ovarian and colorectal cancer cells. By employing whole-genome screens, they identified how cancer cells evade the drug’s effects. Surprisingly, the research found that cancer cells did not adapt their autophagy mechanisms as expected. Instead, they survived by altering their metabolism and division pathways.
“We thought the main interaction of hydroxychloroquine with cancer was through autophagy, but it appears that processes unrelated to autophagy may be more crucial for cancer cell survival,” Delaney stated.
Toward Innovative Combination Therapies
These insights pave the way for new combination therapies. Delaney’s team aims to identify additional drugs that can be administered alongside hydroxychloroquine to combat resistance. By combining hydroxychloroquine with treatments targeting cell division, metabolism, or export pathways, researchers hope to enhance therapeutic effectiveness.
Moreover, the findings suggest that hydroxychloroquine could be particularly effective in treating cancers already affected by defects in these newly identified pathways. Conversely, patients without these defects might benefit more from alternative treatments.
“We want to understand which patients would benefit most from these trials,” Delaney emphasized, indicating a shift toward personalized treatment approaches.
In conclusion, the research from the Delaney Lab sheds light on how repurposed drugs like hydroxychloroquine can be harnessed more effectively in the battle against cancer. By uncovering the unexpected ways cancer cells resist treatment, scientists are one step closer to developing robust combination therapies that could improve patient outcomes.
Further Reading: Silvia G Vaena et al, Autophagy unrelated transcriptional mechanisms of hydroxychloroquine resistance revealed by integrated multi-omics of evolved cancer cells, Cell Cycle (2024). DOI: 10.1080/15384101.2024.2402191.