Singapore, August 2024 – Researchers from Singapore General Hospital (SGH), A*STAR’s Genome Institute of Singapore (GIS), and the Yong Loo Lin School of Medicine at the National University of Singapore (NUS Medicine) have identified a new clade of Candida auris, increasing the total number of known clades to six. This breakthrough has significant implications for global public health.
Candida auris, a highly transmissible and difficult-to-eradicate fungus, primarily affects patients with severe underlying conditions. Those with invasive medical devices such as breathing or feeding tubes, as well as catheters, are particularly vulnerable to infections ranging from superficial to severe and life-threatening.
The World Health Organization (WHO) recognized C. auris as a critical priority for research and public health action in 2022. Additionally, the U.S. Centers for Disease Control and Prevention (CDC) has classified it as an ‘urgent antimicrobial resistance threat,’ underscoring the urgent need to address this growing public health challenge.
Dr. Karrie Ko, co-first author of the study and Consultant in the Department of Microbiology at SGH, emphasized the importance of enhancing surveillance capabilities in light of this discovery. “The implications of this discovery extend far beyond the laboratory. With the identification of the sixth Candida auris clade, there is a pressing need to improve surveillance and develop new strategies to monitor and control its spread,” Dr. Ko stated. She is also the Genomics Director of the Pathology Academic Clinical Program under SingHealth and Duke-NUS Medical School.
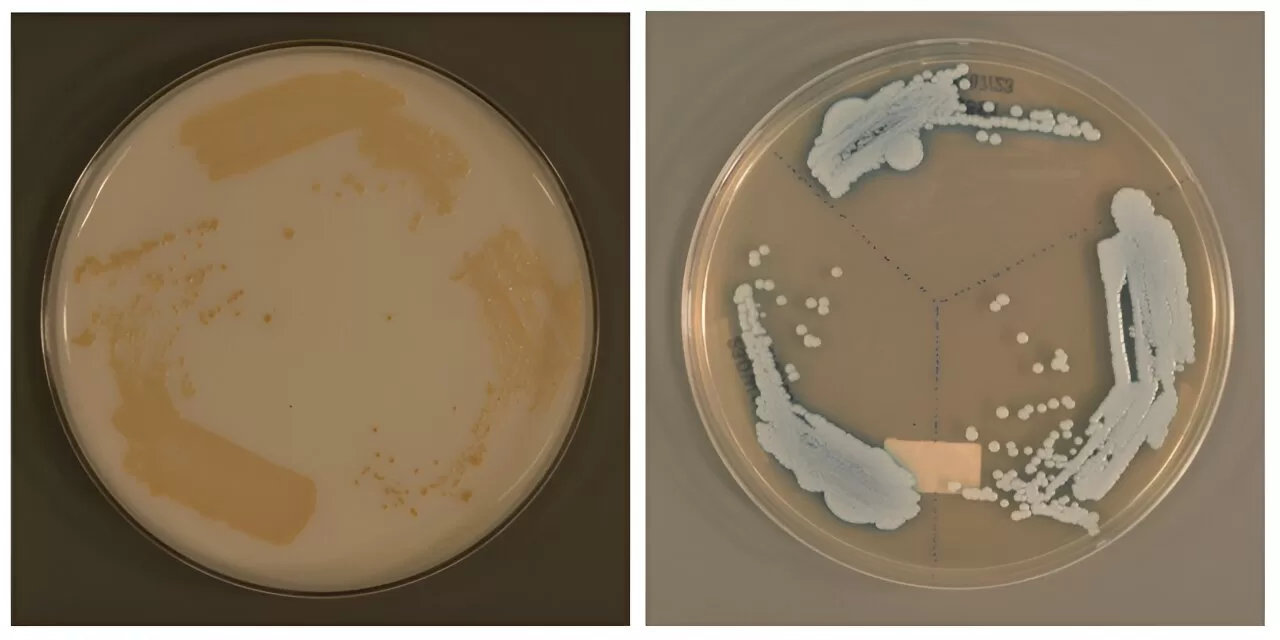
The new clade was detected in 2023 after an SGH patient, who had not traveled abroad in two years, tested positive for C. auris. This prompted further investigations, leading to the identification of the novel clade. A*STAR’s GIS employed a machine-learning technique to track potential new C. auris clades, reconstructing and characterizing the fungal genomes to confirm the emergence of this new variant.
The research team discovered that the patient carried C. auris from a clade distinct from the previously identified five. An examination of historical patient records revealed two additional cases of this new clade. Their findings were published in “Detection and Characterization of a Sixth Candida auris Clade in Singapore: A Genomic and Phenotypic Study” in The Lancet Microbe in July 2024.
SGH has implemented an active surveillance program to monitor high-risk patients for C. auris upon admission. Patients who test positive are isolated, and additional screenings are conducted for those in close proximity to contain potential outbreaks.
Dr. Chayaporn Suphavilai, co-first author and Senior Scientist at A*STAR’s GIS, highlighted the potential of machine learning in enhancing surveillance. “Our study demonstrates that machine learning can improve surveillance by detecting unusual outlier genomes. This workflow enables continuous learning from new data, enhancing our ability to identify and investigate novel genomes promptly,” Dr. Suphavilai said.
Associate Professor Niranjan Nagarajan, senior author of the study and Associate Director of Genome Architecture at A*STAR’s GIS, emphasized the critical role of genomic surveillance. “Integrating genomics, metagenomics, and collaborative research is essential for understanding emerging pathogens and enhancing our preparedness for public health threats.”
For further reading, see Chayaporn Suphavilai et al, “Detection and Characterisation of a Sixth Candida auris Clade in Singapore: A Genomic and Phenotypic Study,” The Lancet Microbe (2024). DOI: 10.1016/S2666-5247(24)00101-0.