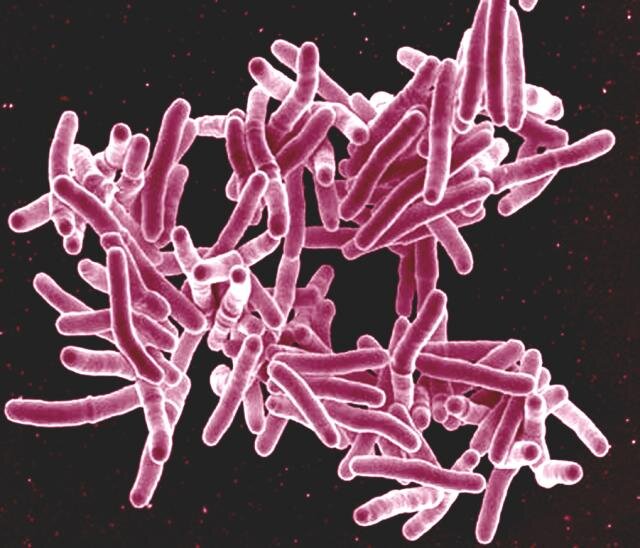
Tuberculosis (TB) remains a major global health challenge, infecting over 10 million people in 2022 alone. The airborne pathogen, Mycobacterium tuberculosis, primarily affects the lungs, leading to symptoms such as chronic cough, chest pain, fever, fatigue, and weight loss. While TB is more prevalent in certain regions, a recent outbreak in Kansas has led to two deaths, marking one of the largest recorded outbreaks in the United States.
Historically, TB has been treated with antibiotics, but the increasing prevalence of drug-resistant strains has intensified the urgency for new treatment options. In a groundbreaking study published in the Proceedings of the National Academy of Sciences, researchers have leveraged artificial intelligence (AI) to expedite the search for antimicrobial compounds that could lead to novel TB drug treatments.
The study was conducted by scientists from the University of California San Diego, Linnaeus Bioscience Inc., and the Center for Global Infectious Disease Research at the Seattle Children’s Research Institute. Linnaeus Bioscience, a San Diego-based biotechnology firm, has been developing a bacterial cytological profiling (BCP) method that provides a rapid way to understand antibiotic mechanisms.
Traditional TB drug discovery has been a slow and labor-intensive process due to the complexity of understanding how drugs interact with Mycobacterium tuberculosis. The newly developed “MycoBCP” technology adapts BCP using deep learning, an advanced AI technique that mimics neural networks in the human brain. This approach allows researchers to analyze TB cell images more efficiently, detecting subtle patterns and structures that were previously difficult to interpret.
“This is the first time machine learning and AI have been applied in this way to bacteria,” said co-author Joe Pogliano, a professor in the Department of Molecular Biology at UC San Diego. “Tuberculosis images are inherently difficult to interpret by the human eye and traditional lab methods. Machine learning provides a much more sensitive way to detect differences in cell structures.”
The MycoBCP technology was developed over two years, with study lead authors Diana Quach and Joseph Sugie training AI tools on more than 46,000 TB cell images. Both researchers, now at Linnaeus Bioscience, earned their Ph.D.s from the Shu Chien-Gene Lay Department of Bioengineering and completed postdoctoral work in Pogliano’s lab.
“TB cells tend to clump together, making it difficult to define cell boundaries,” said Sugie, who serves as chief technology officer at Linnaeus Bioscience. “Instead of trying to manually identify these boundaries, we allowed AI to recognize patterns within the images.”
Linnaeus collaborated with TB expert Tanya Parish of the Seattle Children’s Research Institute to adapt BCP for mycobacteria. This cutting-edge system has significantly accelerated the process of identifying potential drug candidates, streamlining the ability to determine how new compounds affect TB bacteria.
“A crucial step in drug development is understanding how a compound works, which has traditionally been challenging and time-consuming,” said Parish, a co-author of the study. “This technology enables us to prioritize molecules based on their mode of action, allowing us to focus on the most promising candidates. We were excited to work with Linnaeus on advancing this approach for M. tuberculosis.”
The research team, which includes Marc Sharp, Sara Ahmed, Lauren Ames, Amala Bhagwat, and Aditi Deshpande, believes this AI-driven approach will help overcome the hurdles of TB drug resistance, paving the way for more effective treatments in the future.
Reference: Sugie, Joseph et al. “Deep learning–driven bacterial cytological profiling to determine antimicrobial mechanisms in Mycobacterium tuberculosis.” Proceedings of the National Academy of Sciences (2025). DOI: 10.1073/pnas.2419813122.
Disclaimer: This article is based on a recent study and is intended for informational purposes only. It does not constitute medical advice or endorsement of specific treatments.