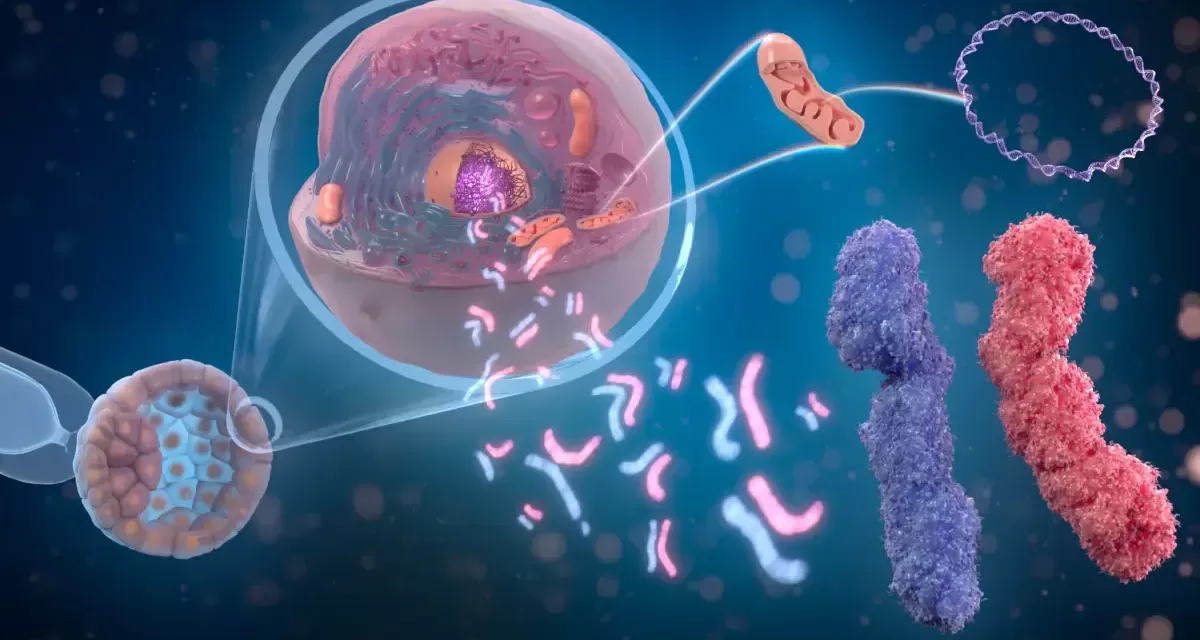
A groundbreaking new resource detailing the functions of more than 20,000 human genes has been released, marking a significant milestone in biomedical research. Developed by the Gene Ontology Consortium, this extensive database—termed the “PAN-GO functionome”—integrates large-scale evolutionary modeling with experimental data to provide the most comprehensive and accurate depiction of human gene functions to date.
For the first time, scientists from the Keck School of Medicine of USC, the Swiss Institute of Bioinformatics, and other institutions have combined genetic data from humans with information collected from various model organisms, such as mice and zebrafish. This approach enables researchers to bridge gaps in direct human gene research by leveraging evolutionary insights.
A New Era in Biomedical Research
The Gene Ontology Consortium has been a cornerstone of biomedical research for over 25 years, assisting scientists in analyzing large-scale “omics” experiments—studies of DNA, RNA, proteins, and other biological molecules. Traditionally, researchers seeking to understand the role of specific genes in disease processes would have to sift through thousands of scientific papers. The PAN-GO functionome streamlines this process by compiling and categorizing gene functions based on the most reliable scientific evidence available.
“Our knowledge base allows scientists to go from just a list of genes to an understanding of their biological functions, including what might be useful for treatment,” explained Dr. Paul D. Thomas, a principal investigator of the Gene Ontology Consortium and professor at the Keck School of Medicine.
Evolutionary Insights Drive Discoveries
The PAN-GO functionome is unique in that it incorporates evolutionary modeling to enhance our understanding of gene functions. By tracking the evolutionary history of thousands of genes and their related proteins, researchers can infer gene functions even when direct human experimental data is lacking.
“We’ve amassed an authoritative reference on human gene functions, and by adding evolutionary context, we’ve created a more complete, accurate, and concise description of the functions encoded by human genes,” Thomas added.
The resource was compiled by a global team of over 150 biologists who meticulously reviewed more than 175,000 scientific publications on gene function. Each gene has been categorized according to its role in biological processes such as cell division, immune response, and molecular transport.
A Call to Action for Researchers
The Gene Ontology Consortium is encouraging scientists worldwide to utilize the PAN-GO functionome in their research. The information is structured in a machine-readable format, making it compatible with artificial intelligence and computational tools for faster and more effective data analysis.
Moreover, researchers are invited to contribute updates and refinements to the knowledge base, ensuring its continued evolution and accuracy. Although the resource is the most comprehensive of its kind, approximately 18% of human protein-coding genes—around 3,600 in total—still lack experimental data regarding their biological functions.
“We now have a clear picture of where research is needed, and this can help direct future studies to uncover unknown gene functions,” Thomas noted.
Looking Ahead
With the completion of the PAN-GO functionome, biomedical scientists now have an invaluable tool to accelerate discoveries in genetics, disease mechanisms, and potential treatments. By combining cutting-edge evolutionary modeling with decades of research, this resource promises to enhance our understanding of the human genome like never before.
For more details, the full study is available in the journal Nature under DOI: 10.1038/s41586-025-08592-0.
Disclaimer: The information presented in this article is based on current research findings and is subject to further updates as new data becomes available. The PAN-GO functionome is a continuously evolving resource, and its use in research should be complemented with other scientific methods and peer-reviewed studies.