Researchers discover a novel RNA-based regulatory system in gut bacteria, offering new insights for therapeutic strategies
In a groundbreaking study, researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität (JMU) in Würzburg have unveiled a protein and a group of small ribonucleic acids (sRNAs) in Bacteroides thetaiotaomicron, a prominent gut bacterium, that regulate sugar metabolism. This discovery promises to reshape strategies for improving gut health through microbiota modulation. The research, recently published in Nature Communications, opens up new possibilities for therapeutic interventions that leverage the microbiota to promote overall health.
The human gut plays a crucial role in maintaining well-being, with its microbiota—an ecosystem of diverse microorganisms—playing an integral part in digestion, immunity, and metabolic processes. These microbial communities are known to adapt to the ever-changing conditions of the gut environment, a critical factor for their successful colonization. Understanding how gut bacteria adjust their metabolism in response to varying nutrient availability has become a central focus of microbiota research.
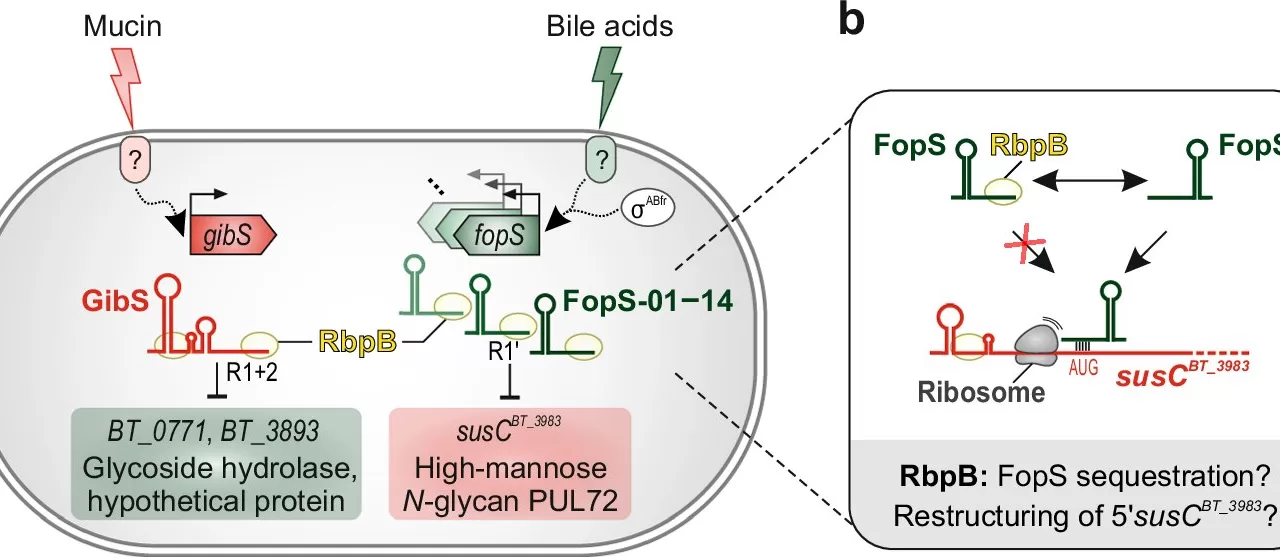
Among the most abundant gut microbes is Bacteroides thetaiotaomicron, a bacterium that thrives in the human gut due to its ability to break down complex carbohydrates. This ability is mediated by a system of proteins called polysaccharide utilization loci (PULs), which enable the bacteria to degrade and import specific polysaccharides. While the production of PUL complexes is known to be tightly regulated at the transcriptional level, the post-transcriptional mechanisms that govern these processes have remained poorly understood—until now.
In a collaborative effort with Vanderbilt University and the University of Toronto, the research team at HIRI and JMU has uncovered a sophisticated RNA-based regulatory system that controls PUL expression in Bacteroides thetaiotaomicron. Their findings suggest that the RNA-binding protein RbpB, along with a family of small non-coding RNAs called FopS, plays a crucial role in regulating the bacterium’s sugar metabolism and adaptation to environmental shifts.
Lead author Ann-Sophie Rüttiger and corresponding author Alexander Westermann emphasized that the absence of RbpB severely impairs the bacterium’s ability to colonize the intestine, highlighting the importance of this protein in gut colonization and nutrient adaptation. The research revealed that RbpB interacts with hundreds of cellular transcripts, including the FopS family of sRNAs, which work together to optimize the bacterium’s catabolic processes in response to fluctuating nutrient conditions.
“This study provides critical insight into how RNA-coordinated metabolic regulation helps microbiota species adapt to their environment,” said Rüttiger. “By understanding these mechanisms, we gain a deeper understanding of the microbiota’s role in human health and its potential therapeutic applications.”
The team plans to continue exploring the structural properties of RbpB and its RNA-binding mechanisms, as well as comparing it with other RNA-binding proteins found in different gut microbiota species. These efforts could reveal shared regulatory hubs that control metabolic processes across a broad range of gut bacteria.
The findings also have significant implications for medical treatments. A more in-depth understanding of bacterial gene and protein functions could pave the way for novel therapies aimed at combating gut-related diseases, such as infections and inflammatory bowel diseases, while promoting the beneficial effects of the gut microbiota.
“Our results open up new avenues for manipulating the microbiota to enhance gut health and overall well-being,” concluded Westermann. “This research holds promise for developing innovative strategies that harness the power of the microbiota to treat and prevent diseases.”
For further reading, see the study: The global RNA-binding protein RbpB is a regulator of polysaccharide utilization in Bacteroides thetaiotaomicron, Nature Communications (2025). DOI: 10.1038/s41467-024-55383-8.