Indian Scientists, in partnership with Scientists from China, Russia and Brazil, will carry out genomic sequencing of SARS-CoV-2 and studies on the epidemiology and mathematical modelling of the COVID-19 pandemic. This will help trace genetic mutations, recombinations as well as distribution of the virus and also make projections about the future of its spread.
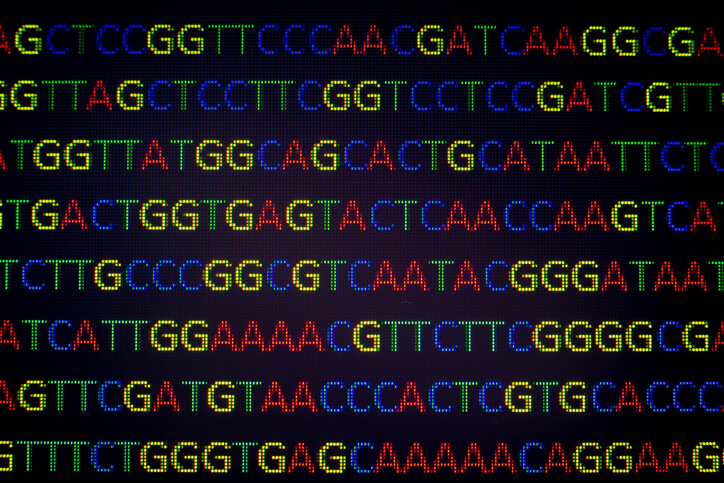
Whole-genome sequencing is required for the identification of genetic mutations and recombinations of the virus, while epidemiological studies can help assess its distribution. Mathematical modelling is required to assess its future spread.
Keeping this in mind, a research plan has been made by including the expertise of scientists and engineers from diverse backgrounds. A consortium consisting of Dr Ch Sasikala, Professor, Centre for Environment, Institute of Science and Technology, Jawaharlal Nehru Technological University Hyderabad, Yuhua Xin, Professorate Senior Engineer Institute of Microbiology, Chinese Academy of Sciences, Beijing, China, Ivan Sobolev, Senior Researcher, Federal Research Center of Fundamental and Translational Medicine, Timakova, Russia, Dr Marilda Mendonça Siqueira, Respiratory Viruses and Measles Laboratory, Oswaldo Cruz Institute, Fiocruz., Rio de Janeiro, Brazil will carry out different arms of this BRICS-Multilateral Research and Development Project.
Under this research supported by the Department of Science and Technology, India and Brazil sides will assess the distribution of SARS-CoV-2 in environmental samples through metagenome analysis for wastewater-based epidemiology (WBE) surveillance. Chinese and Russian scientists will carry out the Real-Time PCR detection of SARS-CoV-2 in biological material (nasopharyngeal swabs) from patients with symptoms of respiratory diseases and investigate the genomic variability, comparative genomics and phylogenetic analysis.
The genomic, metagenomic and epidemiological data from India, China, Russia and Brazil will be integrated to develop mathematical models for mutations analysis, population genetics, phylogenetic relationship, recombination analysis and risk evaluation to reveal the spread network and dynamics of the virus. This can help trace the spread routes and dynamics of the virus. The database developed by the different groups will also compare the distribution and survival of the virus in the different regions and establish the surveillance of the relevant early warning system.
The collaborative research plan has been developed considering the strengths of international collaborators from the Institute of Microbiology, Chinese Academy of Sciences of China, Federal Research Centre of Fundamental and Translational Medicine of Russia and Respiratory Virus and Measles Laboratory, Oswaldo Cruz Institute of Brazil. The study will provide a common platform to share and analyse the data of four different countries and understand the spread routes and transmission dynamics of the virus.
For further details, Dr Ch Sasikala ([email protected]; [email protected]) can be contacted