Hamburg, Germany — A groundbreaking new tool is shedding light on how malaria parasites cling to red blood cells, offering new insights into the disease’s ability to evade the immune system. The research, published today in eLife, introduces a novel method for studying the variable proteins that allow Plasmodium falciparum, the parasite responsible for malaria, to adhere to red blood vessels—a key factor in its ability to cause disease.
Malaria remains one of the deadliest infectious diseases worldwide, with millions of cases each year. The ability of P. falciparum to attach to red blood cells, known as cytoadhesion, is critical for its survival and pathogenicity. This attachment helps the parasite avoid immune detection and leads to the buildup of infected red blood cells in vital organs, resulting in severe, life-threatening complications.
A protein family called “P. falciparum erythrocyte membrane protein 1” (PfEMP1) facilitates cytoadhesion. P. falciparum parasites are capable of producing multiple variants of this protein, and each parasite typically expresses just one type at a time. This ability to switch between different variants of PfEMP1 is one of the parasite’s strategies to evade the immune system. However, studying these variants has been a significant challenge for researchers.
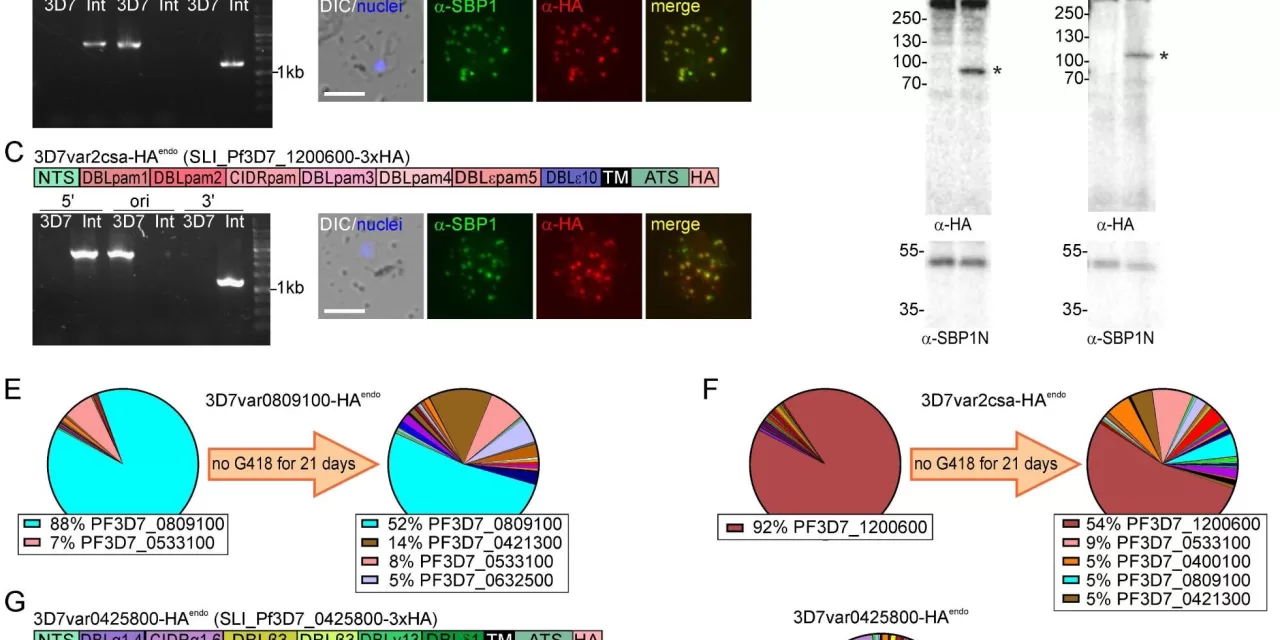
A team of scientists led by Jakob Cronshagen from the Bernhard Nocht Institute for Tropical Medicine in Hamburg, Germany, developed a new technique called “selection linked integration” (SLI) to address this problem. This method allows the generation of parasite lines that exclusively express a single variant of PfEMP1. By enriching these parasites through antibiotic selection, the researchers were able to create a uniform population of parasites, facilitating more detailed studies of how PfEMP1 functions during infection.
“Our method enables us to study individual PfEMP1 variants in greater detail, which has been a longstanding challenge in malaria research,” said Cronshagen, who was a Ph.D. student at the time of the study. “Using SLI, we were able to generate parasite lines expressing a specific tagged version of PfEMP1, allowing us to track its movement and interaction with other proteins.”
The study also explored the “PfEMP1 proxiome”—the group of proteins that work closely with PfEMP1 during malaria infection. In addition to confirming previously known interactions, the researchers identified two new proteins that play a role in the cytoadhesion process, providing fresh targets for future research.
One of the most promising aspects of the study is the ability to mimic the parasite’s natural ability to switch between different PfEMP1 variants. After removing the antibiotic used to select for a specific PfEMP1, the researchers observed that the parasites evolved from a uniform population to a more diverse one, with multiple PfEMP1 variants being expressed. This finding provides a better model for understanding how the parasite adapts during an actual infection.
The new tool could have profound implications for malaria treatment. By allowing researchers to more effectively study and target the proteins responsible for cytoadhesion, the findings may contribute to the development of new therapeutic strategies to combat the disease.
“As we uncover the structural details of how PfEMP1 functions and how it is neutralized by the immune system, we can better understand the complex pathology of malaria,” said senior author Tobias Spielmann, a research group leader at the Bernhard Nocht Institute for Tropical Medicine. “This new approach to generating parasite lines could open up new avenues for blocking these pathogenic proteins, providing potential therapeutic strategies.”
With malaria continuing to pose a significant global health threat, this innovative tool marks a major step forward in the fight against the disease, offering new insights that could lead to more effective treatments.
Source: Jakob Cronshagen et al, “A system for functional studies of the major virulence factor of malaria parasites,” eLife (2024). DOI: 10.7554/eLife.103542.1